Laboratory of Virology and Infectious Disease
A series of crystal structures shows the NS3 helicase at work.
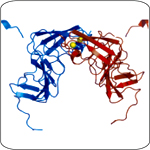
Crystal structure of NS5A domain I shows a groove that may bind RNA.
RNA Replication
HCV and its related viruses replicate entirely in the cytoplasm in close association with intracellular membranes. The virally encoded RNA-dependent RNA polymerase (NS5B) and additional replicase proteins are responsible for amplifying the genomic RNA. We have recently solved the X-ray crystal structures of two enigmatic HCV replicase components. The NS3 helicase is required for RNA amplification, but its purpose is unclear. By capturing a series of molecular "snapshots" we have revealed that this enzyme unwinds nucleic acid with a ratchet-like mechanism. NS5A is a zinc-binding phosphoprotein that plays important but undefined roles in replication and infectious virus assembly. The structure of NS5A domain I revealed a groove potentially involved in RNA binding. Ongoing work is aimed at understanding how the structural features of the NS3 helicase and NS5A contribute to their functions. A detailed molecular understanding of HCV RNA replication will assist in the development of therapeutic drugs targeting this process.
Replication is also dependent on signals within the viral genome itself. Sequences at either end of the viral RNA are known to direct translation and to orchestrate positive and negative strand RNA synthesis. Surprisingly, we have also identified important RNA structures within the polyprotein-coding region. A stem-loop within the core gene, as well as a cruciform element in the NS5B coding sequence, have both been shown to play roles in replication. Interestingly, the NS5B element forms a long-range "kissing-loop" interaction with sequences in the 3′ nontranslated region. These studies highlight the fact that the HCV genome is not only a substrate for replication, but also an integral and dynamic player in this process. Identifying proteins that bind to viral RNA elements may lead to additional targets for replication inhibitors.
Further reading
- Gu M, Rice CM. Three conformational snapshots of the hepatitis C virus NS3 helicase reveal a ratchet translocation mechanism. Proc Natl Acad Sci U S A. 2010 Jan 12;107(2):521-8.
- Tellinghuisen TL, Marcotrigiano J, Rice CM. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature. 2005 May 19;435(7040):374-9.
- McMullan LK, Grakoui A, Evans MJ, Mihalik K, Puig M, Branch AD, Feinstone SM, Rice CM. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc Natl Acad Sci U S A. 2007 Feb 20;104(8):2879-84.
- You S, Rice CM. 3' RNA elements in hepatitis C virus replication: kissing partners and long poly(U). J Virol. 2008 Jan;82(1):184-95.
