Short Chain PIP2 and Kir2.2 Complex
Kir2.2 in complex with PIP2
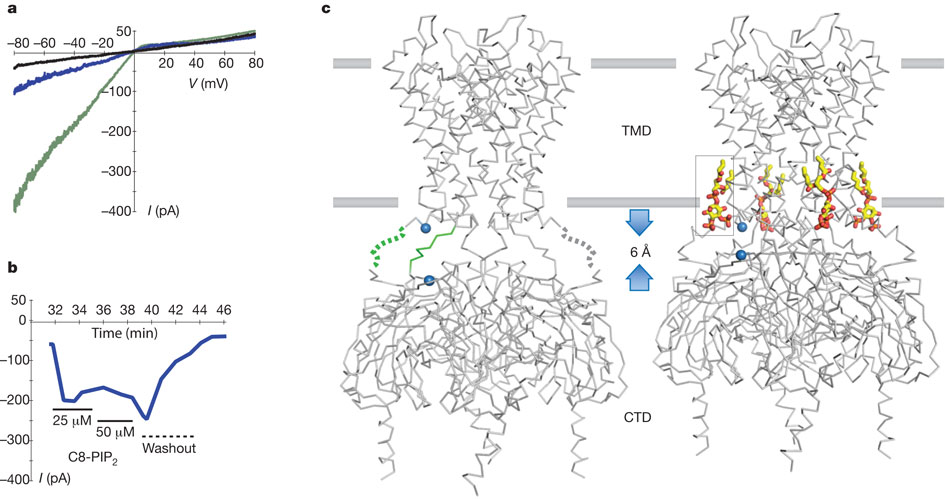
Effect of a short-chain PIP2 on Kir2.2.
a, Endogenous PIP2 depletion causes ‘run down’ of Kir2.2 channels in an excised inside-out patch from Xenopus oocytes as shown by the three macroscopic current traces recorded with a voltage ramp from −80 to +80 mV immediately (green), 30 min (blue) and 50 min (black) after patch excision. b, The short-chain PIP2 added to the bath solution (solid line with concentration indicated below) beginning 32 min after patch excision partially rescued Kir2.2 channel activity. The bath was then perfused (dashed line) at time = 40 min with ~1 ml min−1 bath solution for 3 min. c, X-ray crystal structures of apo- (left, PDB code 3JYC) and PIP2-bound (right, PDB code 3SPI) Kir2.2 tetramer (grey α-carbon traces) viewed from the side with the extracellular solution above. The lipid bilayer boundaries are shown as grey bars. Four PIP2 molecules are shown as sticks and coloured according to atom type: carbon, yellow; phosphorous, orange; and oxygen, red. On PIP2 binding the flexible linker between CTD and TMD consisting of two strands (highlighted green for one subunit, dotted line indicating disordered region in the crystal structure) form helical structures, and the CTD translates towards the TMD by 6 Å. A set of reference atoms (Asp 72 and Lys 220 α-carbons) are highlighted as blue spheres in each structure.
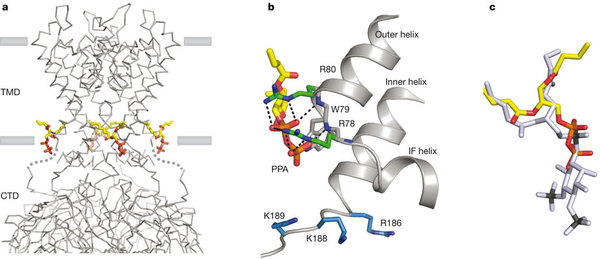
Conserved non-specific lipid binding site in Kirs
a, A grey α-carbon representation of Kir2.2 tetramer in complex with PPA, a small anionic lipid lacking an inositol ring. PPA bound Kir2.2 assumes a closed conformation similar to apo (pdb 3JYC) with the flexible linker elongated and the CTD unengaged. The four PPA molecules are shown as sticks and colored according to atom type: carbon, yellow; phosphorous, orange; and oxygen, red. b, A close-up view of the PPA binding site. PPA contacts Kir at the cytoplasmic end of the outer helix making strong interactions with the guanidiniums of R78 and R80 and the backbone amide nitrogens of the helix turn; similar to the interactions of the 1′ phosphate of PIP2. However, residues (blue sticks) for interacting with the PIP2 inositol 4′,5′ phosphate remain distant to the lipid binding site; R186 orients with its side chain pointing towards the ion conduction pathway. c, Superposition of PPA (colored the same as in panel a) and PIP2 (grey). The position of the 1′ phosphate in PIP2 is between the pyrophosphate of PPA.
Hansen, S.B., Tao, X., MacKinnon, R. (2011). Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature, 477(7365), 495-8. PubMed PMID: 21874019; PubMed Central PMCID – 3324908.
