Nova
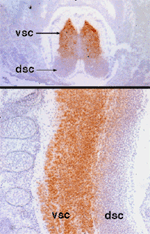
Nova is the target antigen in a disorder associated with defective neuronal inhibition of brainstem and spinal motor neurons, termed POMA (Darnell & Posner, NEJM, 2003). Immunohistochemical and in situ hybridization studies have defined specific expression patterns of these genes in discrete regions of the developing nervous system.
To identify Nova RNA targets, we have utilized RNA selection methods (for method, see for example: Green, R., A. D. Ellington, D. P. Bartel, and J. W. Szostak. 1991. In vitro genetic analysis: selection and amplification of rare functional nucleic acids. Methods (Orlando) 2:75-86.), and validated candidate targets in Nova-null mouse brain ( Jensen KB, Dredge BK, Steffani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YYL, and Darnell RB. Neuron, 25:359-371, 2000. [see minireview Grabowski, Genetic evidence for a Nova regulator of alternative splicing in the brain. Neuron 25:254-6, 2000]; see also more recent validation experiments in Nova KO mice in Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, and Darnell RB. Science, 302;1212-1215, 2003 and Dredge KB and Darnell RB. Mol Cell Biol., 23:4687-4700, 2003.).
X-Ray crystallographic studies have been used to explore the nature of the Nova:RNA interactions. RNA selection was used to produce an aptamer suitable for co-crystallization with Nova KH3 (Jensen KB, Musunuru K, Lewis HA, Burley SK. and Darnell RB. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova KH3 domain. Proc Natl Acad Sci, USA, 97, 5740-5745, 2000.). These crystals revealed that Nova interacts with RNA targets by folding to form a binding pocket capable of coordinating RNA residues in a manner analogous to a second strand of nucleic acid-in essense, the KH domain forms a pseudo-Watson-Crick interaction to specify Nova binding to a specific sequence ( Lewis H, Musunuru K, Jensen KB, Carme E, Chen H, Darnell RB and Burley SK. Cell, 100:323-332, 2000). This mechanism of sequence-specific RNA binding has now been observed for several other KH-type RNA binding proteins (Ramos A, Hollingworth D, Pastore A. RNA. 2003 Mar;9(3):293-8.)
The identification of Nova RNA targets have led us to test predictions about Nova function in Nova KO mice. Several targets have been studied in detail, including pre-mRNA targets encoding the inhibitory glycine (Jensen KB, Dredge BK, Steffani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YYL, and Darnell RB. Neuron, 25:359-371, 2000. [see minireview Grabowski, Neuron 25:254-6, 2000]) and GABAA (Jensen KB, Dredge BK, Steffani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YYL, and Darnell RB. Neuron, 25:359-371, 2000. [see minireview Grabowski, Neuron 25:254-6, 2000] and Dredge KB and Darnell RB. Mol Cell Biol., 23:4687-4700, 2003) receptors. In both instances, Nova regulates exon inclusion, acting as an intronic splice enhancer. We have recently found that Nova can also regulate exon skipping, acting as an exonic splicing silencer (Dredge BK, Stefani G, Engelhard CC, Darnell RB. EMBO J. 2005 Apr 20;24(8):1608-20). Ongoing studies are addressing the mechanism by which Nova proteins regulate these and other splicing targets. Underscoring the significance of these observations, identification of Nova CLIP RNA targets led us to predict changes in a newly identified inhibitory response to LTP stimuli studied by our collaborators Lily and Yuh Nung Jan and colleagues. This inhibitory response involves proteins encoded by Nova target RNAs (GIRK and GABA-B) and is lost in Nova KO neurons (Huang CS, Shi SH, Ule J, Ruggiu M, Barker LA, Darnell RB, Jan YN, Jan LY. Cell. 2005 Oct 7;123(1): 105-18). Ongoing studies are addressing the mechanism by which Nova proteins regulate these and other splicing targets.
Nova CLIP experiments (Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, and Darnell RB. Science, 302;1212-1215, 2003.) has allowed us to build on our identification of Nova as the first mammalian tissue-specific splicing factor (Jensen KB, Dredge BK, Steffani G, Zhong R, Buckanovich RJ, Okano HJ, Yang YYL, and Darnell RB. Neuron, 25:359-371, 2000. [see minireview Grabowski, Neuron 25:254-6, 2000]). Our data suggests that the neuron-specific protein Nova is the predominant (and perhaps only) factor necessary for mediating neuron-specific alternative splicing of a number of very interesting RNAs. Moreover, the method is yielding a large panel of Nova RNA targets, and these are linked biologically. For example, as significant subset encode proteins that function in the synapse, which, taken together with our earlier findings with FMRP, suggest that RNA binding proteins in the brain regulate a biologically coherent set of targets (not a random set).
These observations have been dramatically underscored by recent work in which we have used a new Affymetrix splicing chip to analyze genome-wide alternative splicing changes in Nova Ko and WT brain. The studies confirm in greater detail the finding that Nova specifically regulates exon inclusion/exclusion in a subset of brain transcripts encoding synaptic proteins (Ule J, Ule A, Spencer J, Williams A, Hu J-S, Cline M, Wang H, Clark T, Fraser C, Ruggiu M, Zeeberg BR, Kane D, Weinstein JN, Blume J, Darnell RB. Nat Genet. 2005 Aug;37(8):844-52); this work and its implications in neuronal biology have recently been reviewed (Ule J & Darnell RB. Curr Opin Neurobiol 2006 Jan 13;[Epub]).