Light signal transduction
Light signal transduction pathways are central to the regulation of plant development. They enable information regarding the intensity and duration of specific wavelengths of light to be amplified and coordinated, resulting in complex physiological and developmental responses throughout the life cycle (e.g. germination, seedling de-etiolation, neighbor avoidance and flowering). Plants perceive discrete wavelengths of light through photoreceptors, such as the red/far-red light-sensing phytochromes and the blue light-sensing cryptochromes.
The dramatic switch from skotomorphogenic (etiolated) seedling development to photomorphogenic (de-etiolated) development provides an excellent system to elucidate light signal transduction cascades in plants. Genetic screens of mutagenized seedling populations have identified both positively acting components (receptors, signaling intermediates) and repressors of photomorphogenesis. Our laboratory has focused in particular on identifying intermediates that promote phytochome A signal transduction. Mutant screens identified phytochrome A signal transmission (pat) and long after far-red (laf) mutants. Characterization of these mutants has enabled identification of novel intermediates required for complete responsiveness to activated phytochrome A.
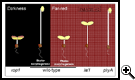 Reproduced, with permission, from Genes and Development Vol 15 (14). © 2001 by Cold Spring Harbor Laboratory Press.
Reproduced, with permission, from Genes and Development Vol 15 (14). © 2001 by Cold Spring Harbor Laboratory Press.Others have identified four biochemical entities: COP1, the CSN, COP10 and DET1 that repress photomorphogenesis in darkness. Characterization of COP1, the CSN and COP10 strongly suggests that ubiquitin-mediated proteolysis plays a critical role in the elimination of signaling components that enable skotomorphogenesis so that they can be replaced by a signaling network that promotes photomorphogenesis. However, the phenotypes of mutants defective in COP gene products indicate that they transduce not only signals from multiple photoreceptors but apparently play central roles in the transduction of signals not directly related to the perception of light.
COP1 is one of approximately 400 Arabidopsis proteins that bear a RING motif. This arrangement of eight cysteine and histidine residues that coordinate two zinc ions is a distinctive feature of a class of E3 proteins. We were the first to demonstrate E3 activity of COP1 and have shown that COP1 plays an important role in phytochrome A signal desensitization via degradation of LAF1, a transcription factor that is required for full response capacity to activated phytochrome A. The demonstration by others that COP1 interacts with a negative repressor of phytochrome A named SPA1 prompted us to test the effect of SPA1 on COP1-mediated ubiquitination. The coiled-coil domain of SPA1 promotes LAF1 ubiquitination, but only at low COP1 concentrations. Based on current insight into the role of nuclear depletion of COP1 after irradiation, this observation suggests a mechanism whereby COP1 in de-etiolated seedlings might act selectively on different substrates to ensure the attenuation of light signals transduced from a specific photoreceptor.
References:
Zhou Q, Hare PD, Yang SW, Zeidler M, Huang L-F, Chua, N-H (2005) FHL is required for full phytochrome A signaling and shares overlapping functions with FHY1. Plant J 43: 356-370
Jang I-C, Yang J-Y, Seo HS, Chua N-H (2005) HFR1 is targeted by COP1 E3 ligase for post-translational proteolysis during phytochrome A signaling. Genes Dev 19:593-602
Zeidler M, Zhou Q, Sarda, Yau C-P, Chua N-H (2004) The nuclear localization signal and the C-terminal region of FHY1 are required for transmission of phytochrome A signals. Plant J 40:355-36
Seo HS, Watanabe E, Tokutomi S, Nagatani A, Chua NH (2004) Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling. Genes Dev 18:617-622
Seo HS, Yang JY, Ishikawa M, Bolle C, Ballesteros ML, Chua NH (2003) LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1. Nature 423:995-999
Moller SG, Kim YS, Kunkel T, Chua NH (2003) PP7 is a positive regulator of blue light signaling in Arabidopsis. Plant Cell 15:1111-1119
Hare PD, Moller SG, Huang L-F, Chua NH (2003) LAF3, a novel factor required for normal phytochrome A signaling. Plant Physiol 133: 1592-1604
Ballesteros ML, Bolle C, Lois LM, Moore JM, Vielle-Calzada JP, Grossniklaus U, Chua NH (2001) LAF1, a MYB transcription activator for phytochrome A signaling. Genes Dev 15: 2613-2625
Moller SG, Kunkel T, Chua NH (2001) A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev 15:90-103
Zeidler M, Bolle C, Chua NH (2001) The phytochrome a specific signaling component PAT3 is a positive regulator of arabidopsis photomorphogenesis. Plant Cell Physiol 42:1193-1200
Bolle C, Koncz C, Chua NH (2000) PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev 14:1269-1278
