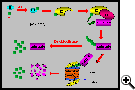
Ubiquitin-Mediated Proteolysis
Ubiquitin is a highly conserved 76 amino acid protein capable of being covalently linked to lysine residues in protein targets. A cascade of enzymatic events that covalently tags substrates with polyubiqitin chains most commonly triggers regulated intracellular protein degradation by a multisubunit protease known as the proteasome. However ubiquitination in yeasts and animals also plays essential roles in DNA repair, protein trafficking and transcriptional regulation through mechanisms that do not involve proteolysis.
The revelation that roughly 5% of the Arabidopsis genome (at least 1,300 genes) apparently contributes to the ubiquitin system strongly implicates the central role of regulated proteolysis in constantly modifying the composition of plant cellular signaling networks. Accordingly, together with others, we have recently revealed pivotal roles for the ubiquitin-proteasome pathway in developmental transitions regulated by light and hormones. From a biotechnological perspective, insight into the regulation of plant signal transduction by protein degradation is likely to vastly expand the repertoire of genes at our disposal to manipulate disease and pest resistance, drought tolerance, improved nutritional properties, and other beneficial crop traits.
Ubiquitin is conjugated in three steps catalyzed by enzymes known as E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme) and E3 (ubiquitin-protein ligase). E3's recruit their cognate target(s) and coordinate their conjugation to ubiquitin. They are thus attributed with most of the specificity associated with regulated proteolysis and constitute amongst the largest plant protein families. Their diversity makes the ubiquitin-proteasome pathway possibly the most complex regulatory system in plants. Nonetheless, thus far, few specific substrates have been identified for plant E3's and there is little insight into how ubiquitination is regulated or the extent to which substrate specificity is maintained. Assuming that accessory proteins associated with E2/E3/substrate complexes fine-tune ubiquitination reactions, the full complexity of the plant ubiquitination apparatus may currently be underestimated.
The Arabidopsis genome also encodes several de-ubiquitinating enzymes. These cysteine proteases cleave isopeptide linkages between either ubiquitins, or between the target and ubiquitin. They contribute another level of control of ubiquitin-mediated regulation by selectively reversing ubiquitination and recycling free ubiquitin back into the cellular pool.
References:
Frugis G, Chua NH (2002) Ubiquitin-mediated proteolysis in plant hormone signal transduction. Trends Cell Biol 12:308-311
Hare PD, Seo HS, Yang JY, Chua NH (2003) Modulation of sensitivity and selectivity in plant signaling by proteasomal destabilization. Curr Opin Plant Biol 6:453-462