Kir2.2 Crystal Structure
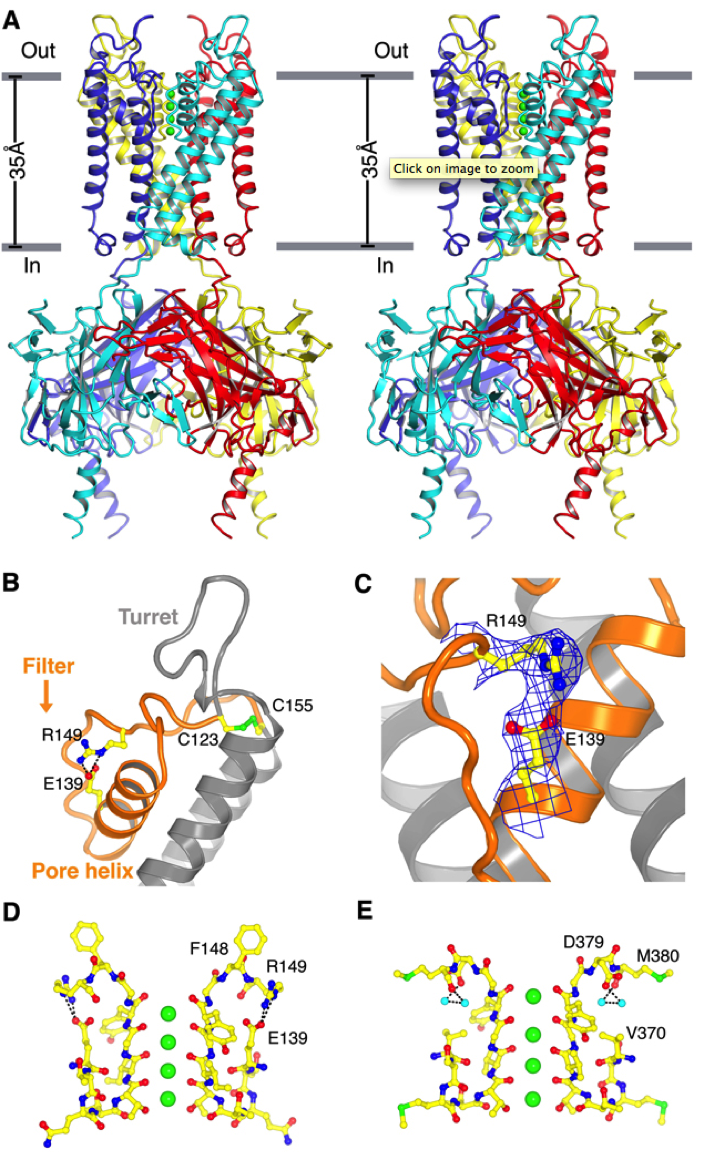
Structure of Kir2.2. (A) Stereoview of a ribbon representation of the Kir2.2 tetramer from the side with the extracellular solution above. Four subunits of the channel are uniquely colored. Approximate boundaries of the lipid bilayer are shown as gray bars. (B) A close-up view of the pore-region of a single subunit (in ribbon representation) with the turret, pore helix and selectivity filter labeled. Side chains of residues E139, R149 and a pair of disulfide-bonded cysteines (C123 and C155) are shown as sticks and colored according to atom type: carbon, yellow; nitrogen, blue; oxygen, red; and sulfur, green. Ionized hydrogen bonds are indicated by dashed black lines. The region flanked by the two disulfide-bonded cysteines is colored orange. (C) Electron density (blue wire mesh, 2Fo-Fc, calculated from 50 to 3.1Å using phases from the final model and contoured at 1.0 σ) is shown for the side chains of E139 and R149 [sticks, colored the same scheme as in (B)] forming a salt bridge. (D and E) K+ selectivity filter of the Kir2.2 channel (D) compared with that of the Kv1.2-Kv2.1 paddle chimera channel [(E), PDB ID 2R9R]. For clarity, only two of the four subunits [sticks, colored with the same scheme as in (B)] are shown. K+ (green spheres), water molecules (cyan spheres), and hydrogen bonds between R149 and E139 (Kir, dashed black lines), or between D379, M380 and waters (Kv, dashed black lines) are shown.
Tao, X., Avalos, J.L., Chen, J., MacKinnon, R. (2009). Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 Å resolution. Science. 326(5960), 1668. doi: 10.1126/science.1180310
