Laboratory of Molecular Neurobiology and Biophysics
Roderick MacKinnon John D. Rockefeller Jr. Professor; Investigator, HHMI

Image created by Yeliz Sezgin
Ion channels catalyze the diffusion of inorganic ions down their electrochemical gradients across cell membranes. Because the ionic movements are passive, ion channels would seem to be extraordinarily simple physical systems, yet they are responsible for electrical signaling in living cells. Among their many functions, ion channels control the pace of the heart, regulate the secretion of hormones into the bloodstream, and generate the electrical impulses underlying information transfer in the nervous system. My research is aimed at understanding the physical and chemical principles underlying ion channel function.
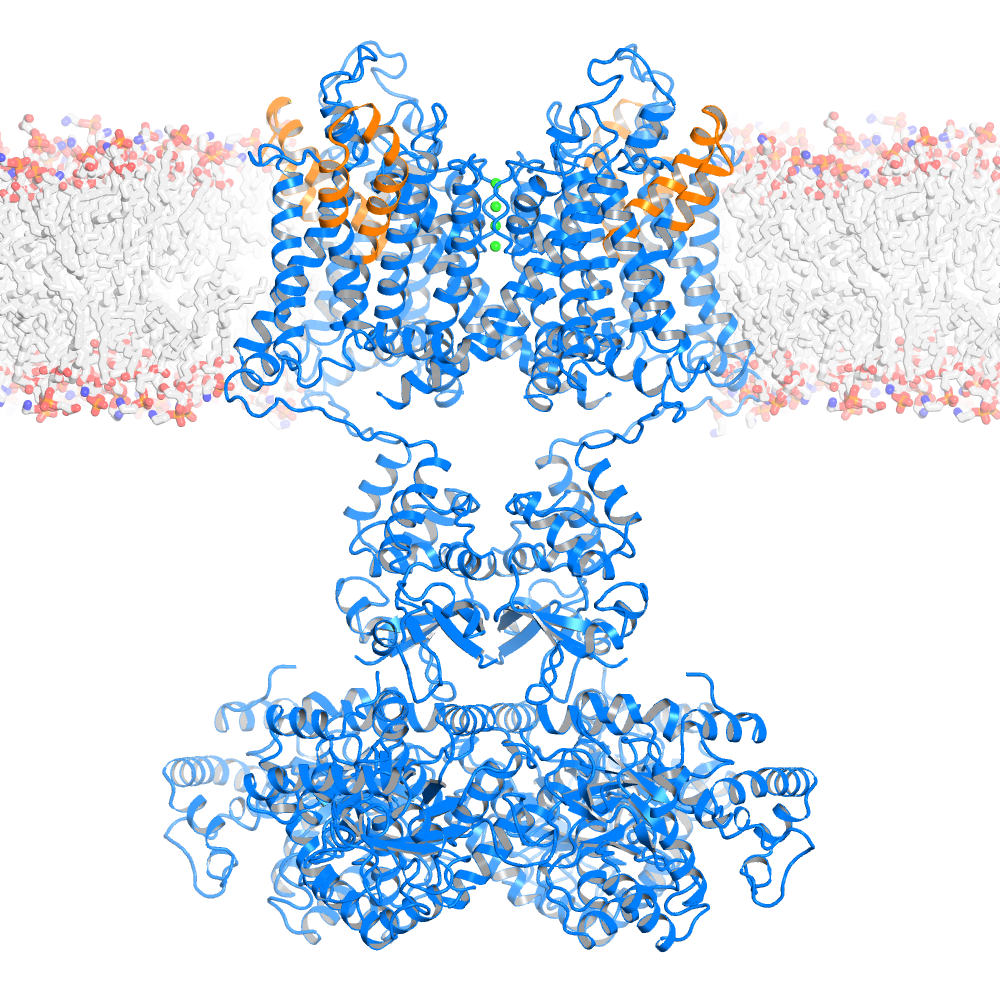
Ribbon representation of the Kv1.2-Kv 2.1 paddle chimera tetramer (PDB 2R9R, voltage sensor paddle colored in orange and the rest of channel in marine, K+ ions shown as green spheres) viewed from the side with the extracellular solution above.
Ion Conduction and Selectivity
Ion channels, like enzymes, have their specific substrates: potassium, sodium, calcium, and chloride channels permit only their namesake ions to diffuse through their pores. Ion selectivity refers to the ability of channels to discriminate among ions. Without ion selectivity, electrical signal production in biology as we know it would be impossible. My laboratory used mutational analysis to show that potassium channels are tetramers of identical subunits and that specific "signature sequence" amino acids are responsible for potassium selectivity. The signature sequence is conserved in all potassium channels throughout nature and forms a structural unit called the selectivity filter.
To understand how the selectivity filter conducts potassium ions rapidly and selectively, we determined the x-ray structures of numerous potassium channels, including the KcsA potassium channel at a resolution higher than 2.0 Å. The selectivity filter is a narrow, 12-Å-long segment of the pore that is lined with carbonyl oxygen atoms, as shown in Figure 1. These atoms act as surrogate water molecules, allowing potassium ions to shed their hydration shell and enter into the pore. We showed that two potassium ions bind in the selectivity filter at once, most likely with a single intervening water molecule. The data support a mechanism in which entry of a third potassium ion on one side of the filter is associated with concerted potassium ion exit from the opposite side, giving rise to efficient potassium ion conduction. Electrostatic repulsion between ions inside the filter prevents the potassium ions from binding too tightly, thus allowing them to conduct at a high rate.
We have developed methods for chemical synthesis and folding of a functional potassium channel in collaboration with Tom Muir (Princeton University). Using these methods, we synthesized potassium channels with main-chain and side-chain chemical groups that do not occur in natural proteins. The chemical synthesis of "unnatural" potassium channels has taught us that a specific conformation of the selectivity filter is important for preventing sodium conduction when potassium ions are not present in solution. When potassium ions are present, they prevent sodium conduction through competition for binding sites.
Chloride channels and transporters mediate the conduction of chloride ions across cell membranes. To understand the chemistry of anion selectivity, we determined the atomic structures of two prokaryotic and one eukaryotic member of the CLC chloride channel family. These anion-selective transport proteins are built on a completely different architectural plan than that of potassium channels, but they nevertheless use many of the same basic physical principles to catalyze selective ionic movement across the membrane. These principles of selective ion movement include oriented alpha-helical-end charges to stabilize ions inside the membrane and short selectivity filters containing multiple ions in a queue to mediate conduction, as shown in Figure 2.These structures have also led us to an atomic model that explains how CLC transporters couple the counter transport of 2 chloride ions per single proton across the cell membrane.
Ion Channel Gating
In humans 78 unique potassium channel genes underlie an impressive level of functional diversity. The different potassium channels have evolved the ability to open – by a process called gating – in response to different forces in their environment. Certain potassium channels are gated by the binding of calcium ions, specific membrane lipids, or G protein subunits. Others respond to mechanical perturbations of the membrane or changes in voltage across the membrane. Thus, the potassium channels control a cell’s electrical state through many different avenues.
To understand the atomic basis of potassium channel gating we have analyzed the function and determined the structures of several different members of the potassium channel family. MthK is a prokaryotic calcium-gated potassium channel, and the BK channel is a eukaryotic cousin of MthK that regulates vascular and airway smooth muscle tone in mammals. Through studies of these channels we have begun to understand how the free energy of calcium binding is converted to mechanical work to open the pore.
Inward rectifier potassium (Kir) channels control the resting membrane voltage in many neuronal and muscle cells. We have recently determined atomic structures to understand how the signaling lipid PIP2 regulates gating in Kir2 (a lipid gated channel) and how G proteins regulate gating in Kir3 (a G protein gated channel). In Kir2, when PIP2 binds to the channel it causes a large cytoplasmic domain to move 6 Å to a position that opens the pore. In essence, PIP2 serves as molecular Velcro to tether two components of the channel together, a requirement to open the conduction pathway. In Kir3, large G protein subunits (b and g subunits) bind to the channel at the intracellular interface where the channel meets the cell’s interior. When the G protein subunits bind they cause a twisting of the a helices that form the pore’s gate, and begin to open it. The structure of Kir3 with the G protein subunits bound complete a story that began in 1921, when Otto Loewi discovered that vagus nerve stimulation slows the heart rate. In Loewi’s experiments, the neurotransmitter acetylcholine was released from the nerve and activated a G protein coupled receptor in the heart pacemaker cells. The G proteins bound to Kir3 and opened it, by a mechanism revealed now in atomic detail, to hyperpolarize the pacemaker cell membrane and slow the heart rate.
Probably the most difficult and in many ways fascinating form of gating we have studied is called voltage-dependent gating. This gating allows "excitable" cells to produce electrical impulses called action potentials: these underlie muscle contraction and nervous system function. We have determined structures of both archaebacterial and eukaryotic voltage-dependent potassium (Kv) channels. In 2005, we published the structure of the Kv1.2 channel from rat, which was the first recombinant eukaryotic membrane protein structure ever solved by x-ray crystallography. Since, we have solved numerous structures to deduce mechanism. Based on our findings we have posited several fundamentally new ideas: (1) voltage-sensor domains are loosely attached to the pore at its perimeter, (2) they contain a helix-turn-helix element that we named the voltage-sensor paddle, (3) the voltage-sensor moves in intimate contact with the membrane, carrying with it charges within the membrane electric field, and (4) the voltage sensor is attached to levers that mechanically open and close the pore. These structures are allowing us to begin to understand how nature, through its ion channel voltage sensors, has evolved the analog of a ‘field effect transistor’ to produce action potentials, which have permitted rapid information transfer in living systems. It is fascinating to realize that voltage-dependent gating, which at a cellular level would seem to be the most complex form of gating, at a molecular level might be the simplest: the voltage across the membrane sets up an electric field that pulls and pushes a charged voltage sensor that is attached to a lever that opens and closes the pore.
Most recently we have begun to study a different form of gating called mechanical gating. We would like to understand how do mechanical forces elicit electrical signals. This form of gating underlies sensory detection as in touch and hearing. It also underlies basic physiological processes such as cell volume regulation and blood pressure control. We and others have observed significant mechanosensitivity in the gating of certain potassium and calcium channels. Now we seek to understand whether mechanosensitivity is physiologically relevant in potassium and calcium channels. We have also determined atomic structures of the human TRAAK channel, which is strongly mechanosensitive, to understand how it transduces mechanical forces, and why the central nervous system expresses a mechanosensitive potassium channel.
The family of potassium channels provides an incredible example of molecular diversity. In an attempt to understand these molecules we never cease to be amazed by life’s simple solutions to what at first appear to be complicated tasks.
A grant from the National Institutes of Health provided partial support for the work on voltage sensing.
