The Mutation Significance Cutoff (MSC)
Links:
- The Mutation Significance Cutoff (MSC) server
- Programs
- The Human Gene Connectome (HGC)
- HGC server (HGCS)
- The Gene Damage Index (GDI)
- GDI server
The Mutation Significance Cutoffs (MSC) of human genes
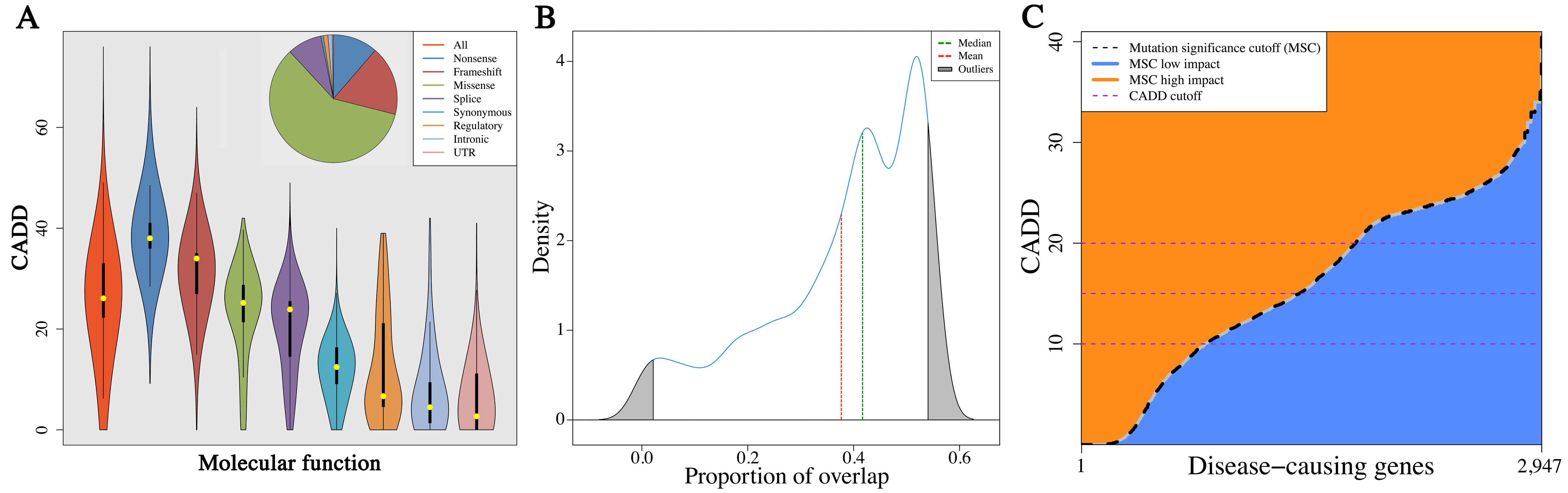
Proposed thresholds of significance for current predictors of biological impact of human genetic variations, such as the CADD, PolyPhen-2 and SIFT scores, are identical for all genes across the genome.
However, the predicted impact scores for known disease-causing mutations in most human genes vary greatly from one gene to another and the scores are strongly associated with the allele frequencies. Therefore, when mutation allele frequencies are taken into consideration, the overall new Human Gene Mutation Database (HGMD) disease-causing allele false negative rates by using the proposed fixed cutoff scores of CADD, PolyPhen-2 and SIFT are >30%.
The Mutation Significance Cutoff (MSC) for each protein-coding human gene is the lower boundary of its 99%, 95%, or 90% confidence interval (CI), generated by either the CADD, PolyPhen-2 or SIFT scores of all its HGMD mutations, and alternatively by ClinVar. The MSC score of a gene represents the lowest expected clinically/biologically relevant CADD cutoff value for that specific gene. For each gene, high phenotypic impact (i.e. possibly damaging) is any CADD/PolyPhen-2/SIFT score equal or above the MSC generated by the specific method, and low phenotypic impact (i.e. benign) is any score below the MSC.
The gene-specific MSC approach reduced the false negative rate to 2%, making it possible for the first time to use variant-prediction methods for WGS/WES hard filtering (removing variants predicted to be benign). We therefore propose that MSC should be used as a first step in filtering out benign variants, whereas other standalone methods would be useful for prioritizing the remaining variants.
The MSC online server is freely available for all non-commercial users.
Citation
When using the MSC method, server and concept, please cite the following paper:
In addition, please cite the method on which your MSC calculation was based:
CADD: PMID 24487276; PolyPhen-2: PMID 20354512; SIFT: PMID 19561590
Contact
In case of problems or questions, please e-mail Yuval Itan at: yitan@rockefeller.edu