Atherosclerosis susceptibility genes in the mouse
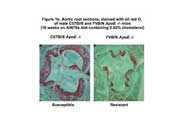
The Breslow laboratory bred the ApoE knockout trait onto different inbred mouse backgrounds and documented major strain differences in atherosclerosis susceptibility (Figures 1a and 1b). Investigators intercrossed atherosclerosis-sensitive, C57Bl/6 (B), and atherosclerosis-resistant strains, FVB/N (F), ApoE knockout mice and used quantitative trait locus mapping of F2 mice to identify a strong susceptibility locus on proximal chromosome 10, Ath11, which was then confirmed in congenic strains. Multiple subcongenic strains revealed Ath11 was complex (Figure 2) with a proximal region (10a) narrowed to 21 genes in females, including the strong candidate Esr1, and a distal region (10b) of 7 genes (Pde7b, Ahi1, Myb, Hbs1l, Aldh8a1, Sgk1, and Raet1) in both genders. Based on aortic microarray expression data, Aldh8a1 is the only one of these genes that is differentially expressed between congenic strains, whereas Myb and Hbs1l are the only genes that contain potential nonsynonymous SNPs. However, there are SNPs present in important regions of other genes as well that could alter their functionality. Additional studies, confirming the presence of these SNPs and determining which are functional and causative in atherosclerosis, will be needed for each of these genes to identify the culprit gene of the 10b region. Further studies are being pursued to determine the culprit genes of the 10a and 10b regions, and to elucidate how the differences between the strains in these genes influence atherosclerosis susceptibility.
Researchers:
Jose Manuel Rodriguez, jrodriguez@rockefeller.edu
Alexandra DeWitt, adewitt@rockefeller.edu